Hey People!!!
So this post is going to be a bit different, no coding, no snapshots, no javascript! I promise!!
This week was the annual bioinformatics conference here in South Africa, it was really cool to see old friends and to know what are they up to these days, and also pretty interesting to know new people and their works.
Submitting a work for this conference was a little different form others, they didn't allow to select between posters or oral presentations. You have to submit your abstract and if accepted they decide the participation type (poster or talk).
Honestly I'm not fan of poster sessions, I found them hard to deal with, people go and check the poster without rally been interested on them, and the presenter find himself repeating the same speech several times in the best case scenario, worst case been nobody show interest in your work and you just stay in front of your poster with nothing to do for as long as the session goes. So, obviously when I wrote my abstract for this conference I was hoping to get an oral presentation.
And... poor me, they select the abstract for a poster... damn! I even though I was not gonna take part of the conference anymore, or just going but not presenting. To be honest, their reasons for not giving me a talk were pretty valid, my abstract didn't have many details of my research, and it was more about Web Visualization in general, and I did it that way because when I wrote it, I didn't have anything I want it to present, all I have where my sketches and a couple of the components I have talk about here, but not real results! Anyway I still thing I could it done a really cool introduction talk about web visualization for bioinformatics.
Anyways, I reckon a poster presentation is better than nothing, so if I was gonna make a poster I was gonna do a really cool one. So I started by asking my self why I dont like posters, and one of the things I hate of poster sessions is that most of the posters look like their author just put the whole content of a paper in the limited space of the poster, so it end up been a lot of text, in a small font that you can't read from distance, a couple of stats graphics, and two or three logos.
It also bothers me the square that research posters are these days, boxes, texts, graphics, logos. But apparently I'm not the only one with this kind of concerns. A friend from the lab sent me this link (http://betterposters.blogspot.com/) after listening to all my quetching!
In contrast to when I wrote the abstract, last week i had a tool in a prototype state that i can really show, so I decide to focus the poster about it, I'm obviously talking about the interaction application that I have chatted so much in this blog (just have a look to this, this and this posts).
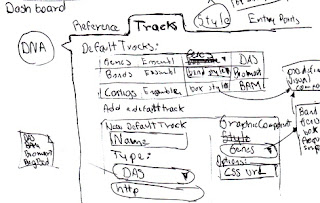
So, with the idea of doing something different and inspired on the application it self, I though I can do the poster resembling the networks that the software is visualizing, and here is a very quick sketch that I did with that idea on mind.
 |
| Sketch for the poster layout |
So the first decision is which software to use to design a poster. A lot of people uses MS PowerPoint for it, but I haven't using that since i met google docs, unfortunately I dont think is feasible to work on design in a web application, specially for something big as a poster, at least not for now.
I have worked with several packages for design before: photoshop, fireworks, illustrator and Gimp. I discover recently that Gimp finally release a version runs on OSX natively, and thats a great deal for me cause I always like it Gimp, but the fact that it was running on X11 made it unnatural to use... Anyway going too technical again, what I'm saying is that if you work in a Mac, you should give it a try to the newest version of Gimp, for you photo editing and design tasks. Is a good software with GNU licence!!
Took me 2 days of work to get the design as I wanted, for one thing it was the first time using this new versin of Gimp, and on the other hand, my design skill were a little rusted. But I did get what I wanted at least in form, the content was still missing. :-)
Besides the normal headers, this layout organize the poster's content in four circles, I took some ideas of the betterposters blog that I mentioned: Using a QR code that points to the website of the tool(actually pointing to this blog), and helping the reader by putting signs of the order that the poster should be read.
Other thing that I considered was which font to use, it really irritates me seen posters, presentation or in fact anything that uses the wrong font, for instance, have you seen the new font that the music channel VH1 is using? I hate it, is too big, looks so unprofessional. Dunno if is just me but AGHHRRHH!
I found a nice idea of how to choose your font between hundreds and is to create a poster with the name of the font using that font, so you can see all the fonts in a simple glance, check the idea here. The fonts they are using in those posters are the over 500 freely available fonts of google, you can use it on web anytime, or download the ones that you like and install it in your computer.
And the content of it, oh well, you can read it yourself, and if you have read my previous blog posts,
you certainly have read over 90% of what's in the poster, only difference is that in the poster I'm a little bit more format, therefore I wont talk about it in this post. You can have a look to a reduce image of the poster:
 |
| Final View of the Poster. |
And a last note, in a different topic, today finally my paper on MyDas is available online, check it out:
http://www.plosone.org/
Time for a beer to celebrate, Cheers!!