Hello There,
Sooner than expected I'm here again writing a new post. To be honest this is some material that I had last week, but the previous post was getting too long and it was taking me a lot of time, therefore I split it and here is a second part. So continuing the idea here I'm gonna show some sketches i made to start having a better understanding of the project.
In this post I will focus on the configuration of a Domain in the Multi-Domain Configuration MDC mode (Remember the 3 parts the project is going to be divided). Here I'm assuming that a lot of components have previously registered in the BIOsual repository, this is important to me because is helping me to define the requirements for the repository mode.
So, once a domain is in the dashboard it needs to be configure, and to open the configuration panel a click in the domain is all am asking for. If is a predefined domain most of the configuration is done, if is a new Domain, the information has to be fill from scratch.
The way that I imagine the domain configuration now, includes 5 different sections, that in the sketches I have organise as tabs: Reference, Tracks, Entry Points, Components and Filters. Obviously at this stage everything can change, but this is the route I'm tracing and as long I reach the goal I'm willing to make changes once on it.
This information can be acquired in different ways.
So in the sketch I'm showing 3 options to load the reference:
Other ways of loading a reference can be using databases, web services, or many other ways that I can not even thing about it and thats why this is based on modules that have to be registered in the BIOsual repository.
So each domain can have tens of tracks, coming from different locations in different formats to be displayed in different ways. Here is another place to modularise.
I will need adaptors for reading formats, I have to get a consensus of which information is relevant here and been able to put it in the same way no matter how is the source defined.
The system also requires to have options to visualise this info, the boxes idea explained above might be generic one, but for instance genes are composed by a series of exons and introns and the common track representation is boxes for the exons and lines for introns. Other annotations have an score, and a histogram representation might be more adequate.
So the sketch number 5 shows that when in the tab to configure the tracks, a list of the already set tracks is displayed, info in the list can be edited, tracks can be removed from the list and the tracks should be draggable to organise its order.
An option to add a new track is presented and if clicked a form should be injected in the config panel. This form depends on the type of source, because the way of access info changes, the form has to be defined as part of the module. So for example if the type of source is DAS the interaction should be similar to the one for the reference.
The visualisation method use for a particular track is also chosen in this form, and it might contain advanced setting for it, such as to associate a CSS style file.
And then, here again the idea of modularise, visual components have to be register to select a region, and is the task of the designer of a MDC to select the appropriate Entry Point selector.
The image shows that for DNA a karyotype selector is set, to allow seen the high level structure of the whole genome.
But more than one Entry Point selector can be desired in a Domain, in the image I also consider that a location selector, and a search in track, the first one can be as simple as text boxes to capture the coordinates, or a slider to select a region or any other interface that allows the selection a region in a genome.
The search in Track is the way I can think in a generic component that can offer the functionality of accessing directly a gene for example.
For now I think that the rule should be define by indicating, the track is going to affect, if is inclusive or exclusive, the parameter to filter, and operator and a value. So in the image, a rule is defined to exclude all the features in Track 1 with a score higher or equal than 0.8
As an optional feature this might not be part of the firs prototypes but I think is something cool to have in mind.
I was getting quite technical for some moment do it might be a boring post, I had fun drawing this sketches, but the most I write the most i realise in what kind of project I'm embarking myself. But i suppose is part of the fun. So if you made it to this part of the post and have any comment/idea/question please let me know.
Un fuerte abrazo,
Gustavo.
Sooner than expected I'm here again writing a new post. To be honest this is some material that I had last week, but the previous post was getting too long and it was taking me a lot of time, therefore I split it and here is a second part. So continuing the idea here I'm gonna show some sketches i made to start having a better understanding of the project.
In this post I will focus on the configuration of a Domain in the Multi-Domain Configuration MDC mode (Remember the 3 parts the project is going to be divided). Here I'm assuming that a lot of components have previously registered in the BIOsual repository, this is important to me because is helping me to define the requirements for the repository mode.
So, once a domain is in the dashboard it needs to be configure, and to open the configuration panel a click in the domain is all am asking for. If is a predefined domain most of the configuration is done, if is a new Domain, the information has to be fill from scratch.
The way that I imagine the domain configuration now, includes 5 different sections, that in the sketches I have organise as tabs: Reference, Tracks, Entry Points, Components and Filters. Obviously at this stage everything can change, but this is the route I'm tracing and as long I reach the goal I'm willing to make changes once on it.
Domain - Reference
Every domain has to have a reference source of information, for instance, the whole genome of an species organised by chromosome for a DNA domain or all the available sequences for the Protein domain.This information can be acquired in different ways.
 |
| 4 - Setting up the reference of a domain. |
- By using a DAS source, this can be done by selection one of the more than 190 DAS reference sources or by directly inputting the URL of the DAS source.
- Sequence Files (web): Some NGS files like BAM allow random access through http requests. This make possible to use a remote file as a reference without having to load the whole file.
The full genome can be divided in several of those files, so the system should allow to load more than one, and provide a way to identify them. Thats why I add the ID field. - Sequence Files(local): Other option might be to have local files as reference. For the case that the files are in the user machine. This might be tricky because of the limited access of javascript into the client machine.
Other ways of loading a reference can be using databases, web services, or many other ways that I can not even thing about it and thats why this is based on modules that have to be registered in the BIOsual repository.
Domain - Tracks
And here guys you will have to apologize me, this is the most confusing of my sketches so far, and that's just a sign that I'm not sure about how to deal with this info.
Tracks are the classical way of displaying biological information in context to a reference (eg. Ensembl, Dasty3, UCSC genome browser, etc.) And basically a track consists in drawing a box in a position that aligns directly with the reference, so you can see where an annotation is located in a chromosome, protein or any other sequence.
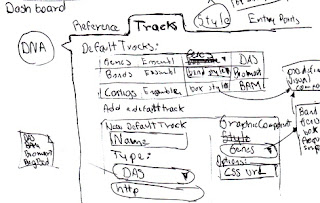 |
| 5 - Setting up the visible tracks of a domain. |
I will need adaptors for reading formats, I have to get a consensus of which information is relevant here and been able to put it in the same way no matter how is the source defined.
The system also requires to have options to visualise this info, the boxes idea explained above might be generic one, but for instance genes are composed by a series of exons and introns and the common track representation is boxes for the exons and lines for introns. Other annotations have an score, and a histogram representation might be more adequate.
So the sketch number 5 shows that when in the tab to configure the tracks, a list of the already set tracks is displayed, info in the list can be edited, tracks can be removed from the list and the tracks should be draggable to organise its order.
An option to add a new track is presented and if clicked a form should be injected in the config panel. This form depends on the type of source, because the way of access info changes, the form has to be defined as part of the module. So for example if the type of source is DAS the interaction should be similar to the one for the reference.
The visualisation method use for a particular track is also chosen in this form, and it might contain advanced setting for it, such as to associate a CSS style file.
Domain - Entry Points
The way of accessing the information vary from domain to domain, for example in genomes, isnormal to go to a particular location indicated by the chromosome and the range of base to visualise, but in proteins there are not chromosomes, and the way usual way to access is by the accession number and the location is not that important because the length of those sequences is not really big.
 |
| 6 - Setting up Entry Points for a Domain |
And then, here again the idea of modularise, visual components have to be register to select a region, and is the task of the designer of a MDC to select the appropriate Entry Point selector.
The image shows that for DNA a karyotype selector is set, to allow seen the high level structure of the whole genome.
But more than one Entry Point selector can be desired in a Domain, in the image I also consider that a location selector, and a search in track, the first one can be as simple as text boxes to capture the coordinates, or a slider to select a region or any other interface that allows the selection a region in a genome.
The search in Track is the way I can think in a generic component that can offer the functionality of accessing directly a gene for example.
Domain - Components
Sorry is getting long again is just 2 more to go, the other tab to configure a domain is to include other components that are not displayed as tracks, mainly because there are not associated with a location or because its content cannot be represented in a unidimensional graphic interface. For instance the 3D structure of a protein or its non positional annotations. |
| 7 - Other Visual Components |
This components have to be also registered in the BIOsual repository, and each of them have to have their own configuration values to be display in this tab.
Domain - Filters
And finally and optional feature that i thing can be really useful, a post query filter based on tracks where the designer can define a rule to accept or reject an of the annotations in the tracks. |
| 8 - Filtering the collected data |
For now I think that the rule should be define by indicating, the track is going to affect, if is inclusive or exclusive, the parameter to filter, and operator and a value. So in the image, a rule is defined to exclude all the features in Track 1 with a score higher or equal than 0.8
As an optional feature this might not be part of the firs prototypes but I think is something cool to have in mind.
I was getting quite technical for some moment do it might be a boring post, I had fun drawing this sketches, but the most I write the most i realise in what kind of project I'm embarking myself. But i suppose is part of the fun. So if you made it to this part of the post and have any comment/idea/question please let me know.
Un fuerte abrazo,
Gustavo.
No comments:
Post a Comment